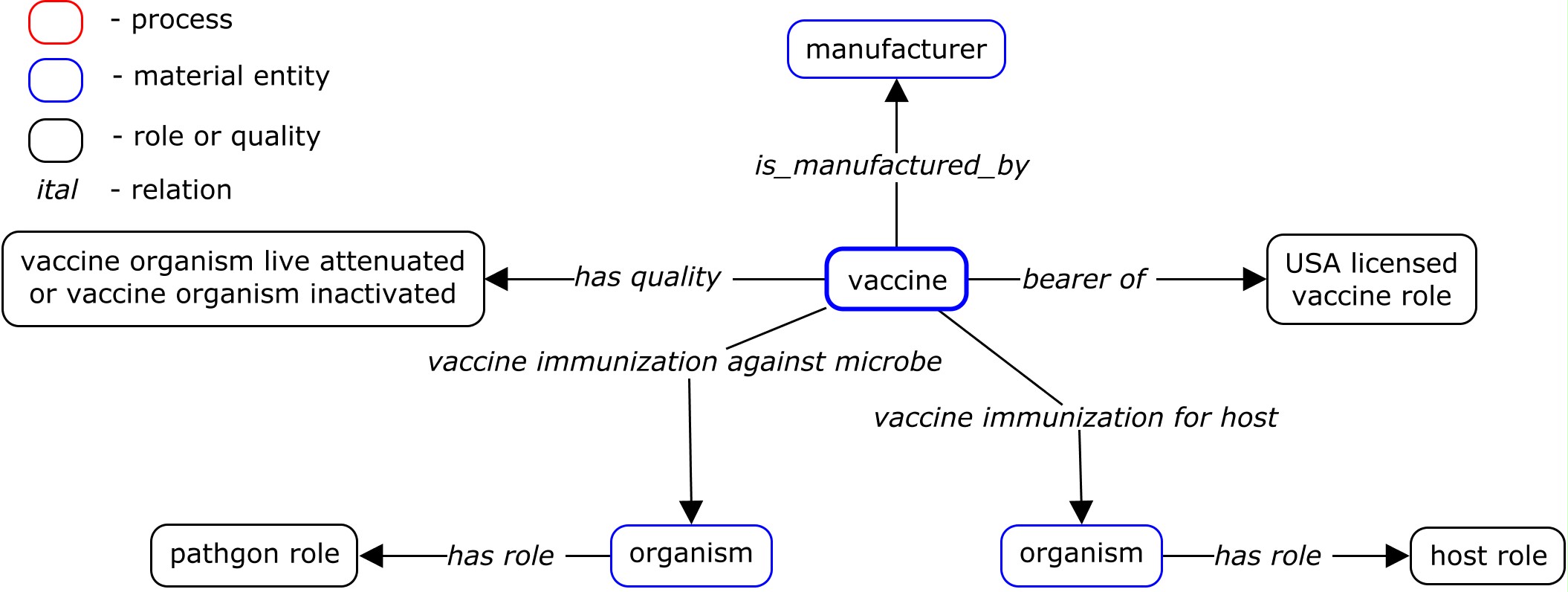
Design Pattern, Template, and Example: VO Vaccine
Task: Generate new VO terms representing a list of USDA-licensed animal vaccines.
This use case study aimed to use Ontorat to generate new VO terms for a list of USDA-licensed animal vaccines. The Ontorat input Excel file and text file contain the same information. The Ontorat can be used to include the following information to the annotations or logical axiom definitions: vaccine name, parent term, host, pathogen, manufacturer, VIOLIN_ID. The vaccine names are the labels of to-be-generated VO classes. Manchester OWL Syntax scripts for expressing the axioms associating the vaccine with other contents are generated and inserted to the Ontorat web forms. It is noted that besides the relations between classes, Ontorat also allows the generation of annotations using annotation properties (e.g., see_Also and term definition). The Ontorat output OWL file can be directly imported to VO.
1. ODP Ontology Design Pattern:
The design pattern for this case is shown by the design pattern diagram below:
2. Ontorat Template Files:
Ontorat setting file: Ontorat_settings_vaccine.txt (.txt format)
Ontorat input template file: Ontorat_input_vaccine_template.xslx (.xlsx format)
3. Ontorat Example:
Introduction: This example aims to add mouse strain terms into the Beta Cell Genomics Ontology (BCGO).
Ontorat input data file (based on template): Ontorat_input_vaccine.xlsx (.xlsx format)
Ontorat output file: Ontorat_output_vaccine.owl (.owl format)
Ontorat output new IDs file when adding new ontology classes: Ontorat_output_newIds_vaccine.xlsx (.xlsx format)
Note: This example is slightly different from the example used in the Ontorat hands-on demo, which is more complex and uses a relatively old version of VO.