Design Pattern, Template, and Example: CLO Cell Line Cells
Task: Add Jappan RIKEN cell lines into Cell Line Ontology (CLO) that indicate the cell line repository as 'RIKEN Cell Bank' with rich annotations including label, alternative term, defintion, term editor, and comments about information of cell line derived from which animal tissue, originators of cell line, etc.
1. Design Pattern:
This use case has been reported in the CLO paper:
Sarntivijai S, Lin Y, Xiang Z, Meehan TF, Diehl AD, Vempati UD, Schürer TC, Pang C, Malone J, Parkinson H, Liu Y, Takatsuki T, Saijo K, Masuya H, Nakamura Y, Brush MH, Haendel MA, Zheng J, Stoeckert CJ, Peters B, Mungall CJ, Carey TE, States DJ, Athey BD, He Y. CLO: The Cell Line Ontology. Journal of Biomedical Semantics. 2014, 5:37. doi:10.1186/2041-1480-5-37.
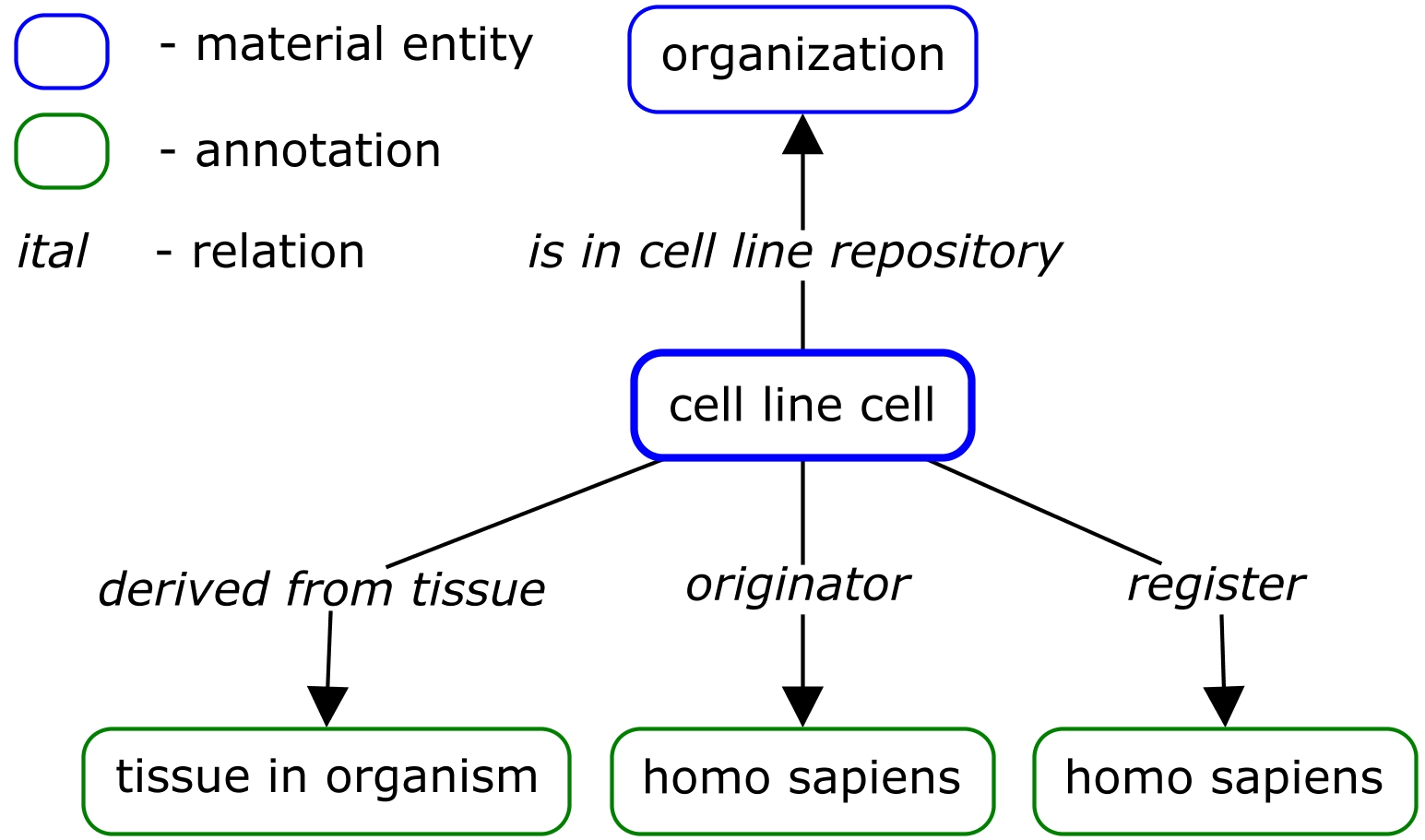
Design pattern diagram:
Notes: The example contains only 10 RIKEN cell lines. Current Ontorat does not support individual. Since 'RIKEN Cell Bank' is an instance, minor modification need to be made of Ontorat output OWL file to change class to individual.
2. Ontorat Template Files:
Ontorat setting file: Ontorat_settings_cellline.txt (.txt format)
Ontorat input template file: Ontorat_input_cellline_template.xlsx (.xlsx format)
3. Ontorat Example:
Introduction: This example aims to add cell line cells based on the CLO. The input data is a list of cell line cells collected from the Japan Rikens institute.
Ontorat input data file (based on template): Ontorat_input_cellline.xlsx (.xlsx format)
Ontorat output file: Ontorat_output_cellline.owl (.owl format)
Ontorat output new IDs file when adding new ontology classes: Ontorat_output_newIds_cellline.xlsx (.xlsx format)
Please feel free to contact us for any questions, comments, and suggestions. Thank you!